Introduction to pmxploit
Patrick Nolain
2019-02-26
pmxploit.RmdThe present article describes briefly how to use pmxploit from R.
Load a NONMEM run archive
run <- load_nm_run(path = "path/to/my_run_archive.tar.gz")Job details
Run information
Model/Data/Files
run$info %>%
bind_cols() %>%
select(run_name, dataset_file, control_stream_file, nodes, nm_version, number_of_subjects, number_of_observations, duration) %>%
mutate(duration = as.character(lubridate::as.period(duration))) %>%
gather(property, value) %>%
kable()| property | value |
|---|---|
| run_name | TMDD_Djebli_al |
| dataset_file | dataset.csv |
| control_stream_file | TMDD_Djebli_al.ctl |
| nodes | 10 |
| nm_version | 7.3.0 |
| number_of_subjects | 527 |
| number_of_observations | 9379 |
| duration | 31612.7990002632S |
Estimation
# Estimation steps summary
run$estimations %>%
map(~ .[c("title", "final_ofv", "minimization", "duration")]) %>%
bind_rows(.id= "n_estimation") %>%
kable()| n_estimation | title | final_ofv | minimization | duration |
|---|---|---|---|---|
| 1: focei | First Order Conditional Estimation with Interaction | 48465.98 | TRUE | 30683.02 |
Model information
Compartments
kable(run$model$compartments)| cmt | name | dv_target | type | unit |
|---|---|---|---|---|
| 1 | DEPOT | FALSE | continuous | NA |
| 2 | Total Alirocumab | TRUE | continuous | NA |
| 3 | Total PCSK9 | TRUE | continuous | NA |
| 4 | PERIPH | FALSE | continuous | NA |
Parameters
| id | type | name |
|---|---|---|
| ETA1 | eta | ETCL |
| ETA2 | eta | EKON |
| ETA3 | eta | EKIN |
| ETA4 | eta | EKDE |
| ETA5 | eta | ETQ |
| ETA6 | eta | ETV1 |
| ETA7 | eta | ETV2 |
| ETA8 | eta | ETKA |
| ETA9 | eta | ETF |
| THETA1 | theta | TVCL |
| THETA2 | theta | TKON |
| THETA3 | theta | TKIN |
| THETA4 | theta | TKDE |
| THETA5 | theta | TVQ |
| THETA6 | theta | TVV1 |
| THETA7 | theta | TVV2 |
| THETA8 | theta | TVKA |
| THETA9 | theta | TVF |
| THETA10 | theta | TVEP |
| THETA11 | theta | TVEA |
| THETA12 | theta | TVLAG |
| THETA13 | theta | COV1 |
Covariates
kable(run$model$covariates)| column | type | name |
|---|---|---|
| ABDO | categorical | ABDO |
| ARM | categorical | ARM |
| DISST | categorical | Disease status |
| EZE | categorical | EZE |
| FIBR | categorical | FIBR |
| ISC | categorical | Administration route |
| SEX | categorical | Gender |
| STATIN | categorical | Statin co-administration |
| STUD | categorical | Study |
| THIGH | categorical | THIGH |
| AGE | continuous | Age (y) |
| BMI | continuous | BMI (kg/m²) |
| BSLDLC | continuous | Baseline LDLC (mg/dL) |
| CLCR | continuous | Creatinine clearance (mL/min) |
| FBSPCSK | continuous | Baseline free PCSK9 (nM) |
| GFR | continuous | Glomerular filtration rate (mL/min) |
| TBSPCSK | continuous | Baseline total PCSK9 (nM) |
| WT | continuous | Weight (kg) |
Diagnostic plots
Spaghetti plot
run %>%
group_by(SEX) %>%
plot_observed_profiles(compartment = 2, y_scale = "log", facetted = FALSE)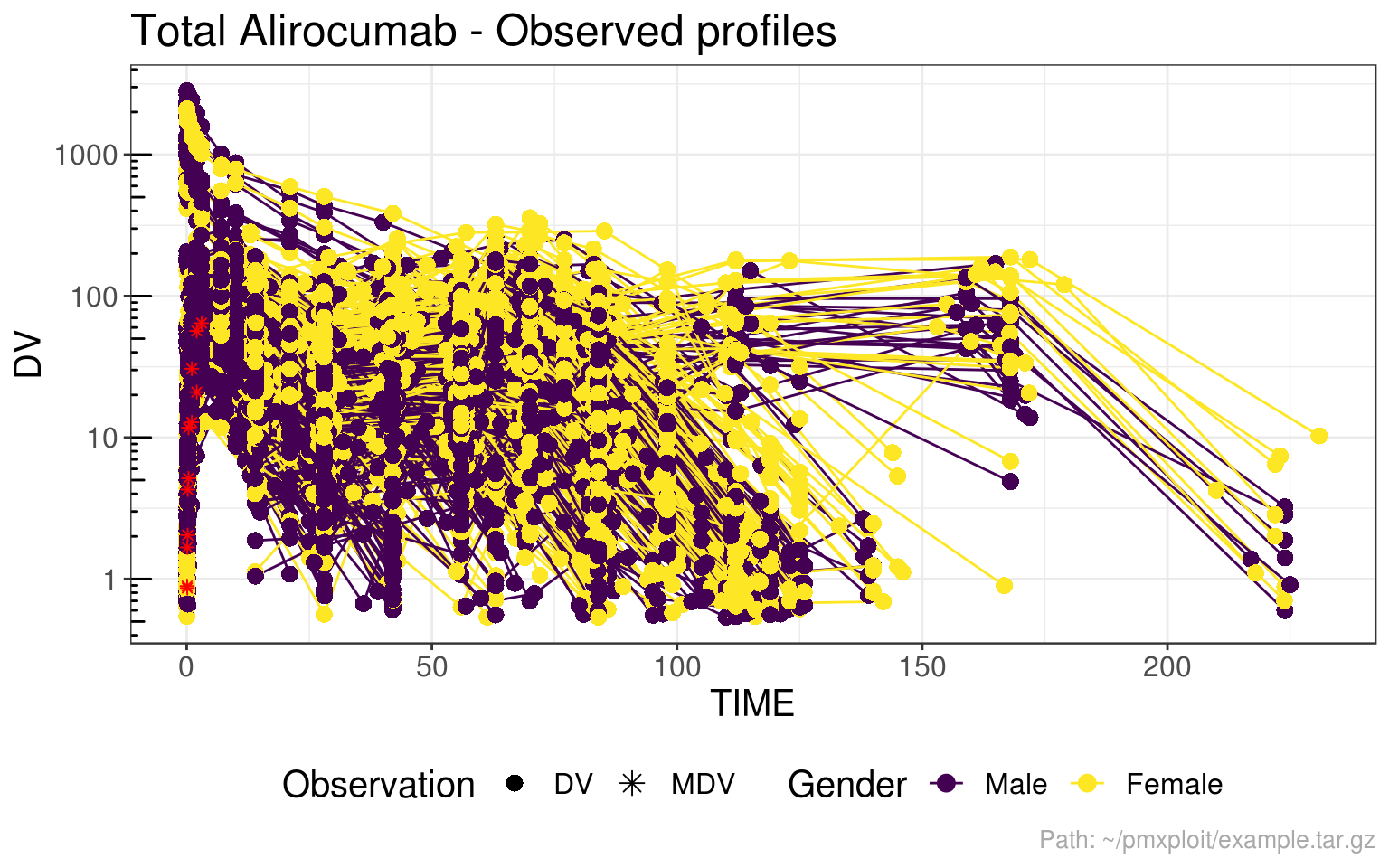
Dependent variable observations vs predictions
plot_dv_vs_predictions(run, compartment = 2, predictions = c("PRED", "IPRED"))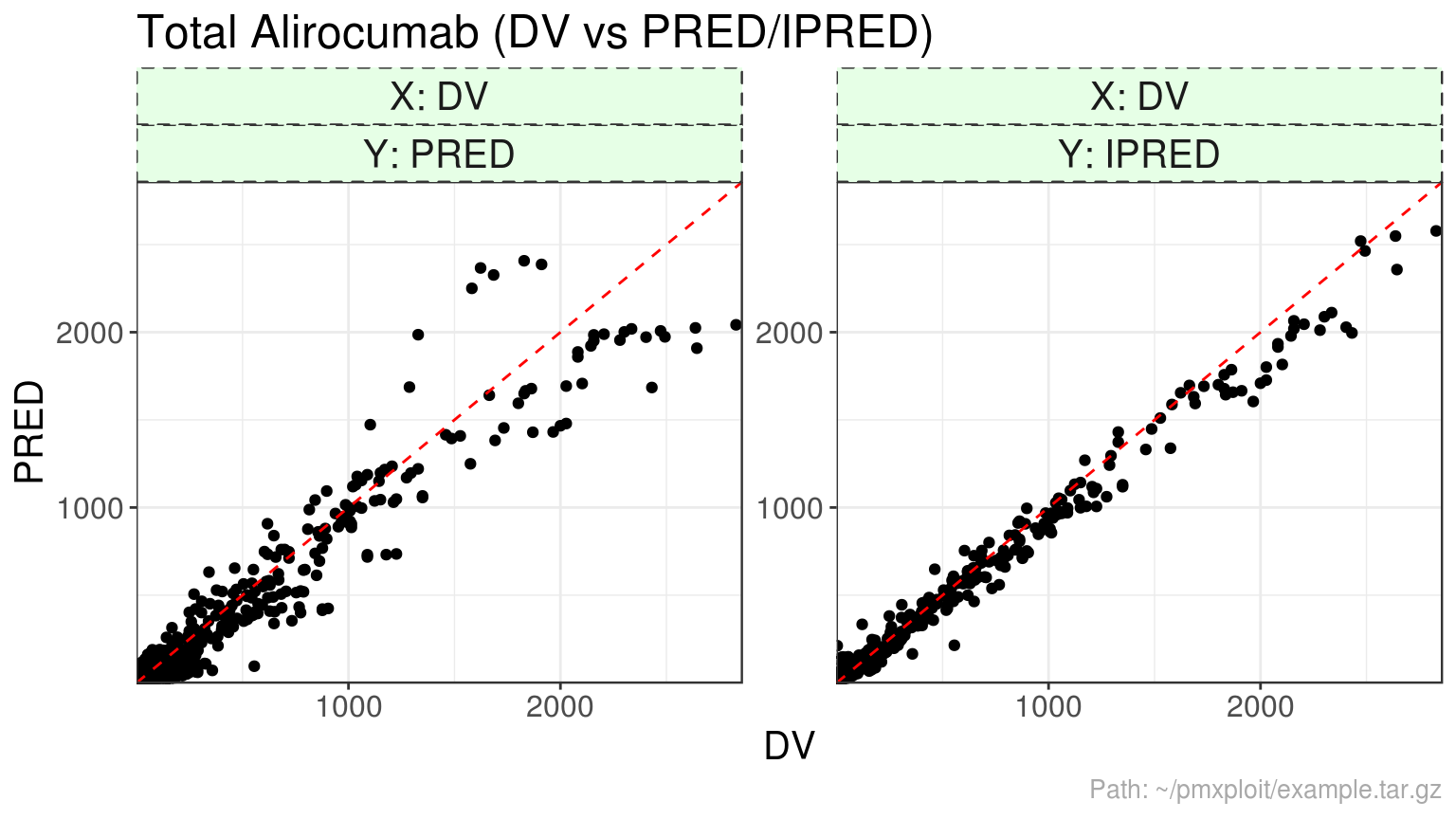
plot_dv_vs_predictions(run, compartment = 2, predictions = c("PRED", "IPRED"), x_scale = "log", y_scale = "log")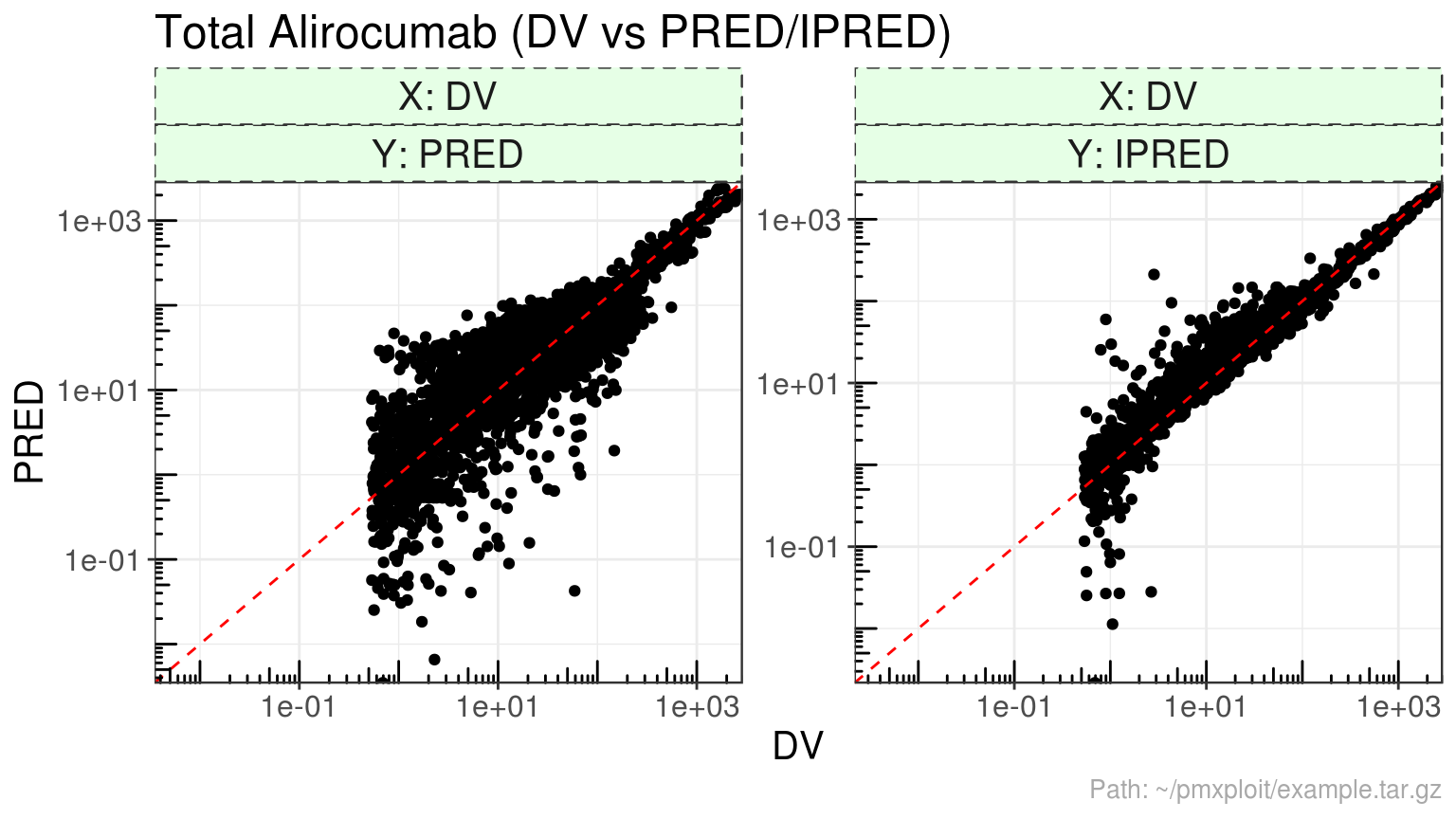
Residuals
plot_residuals(run, compartment = 2, residuals = "CWRES", idv = c("PRED", "TIME"), reference_value = 2)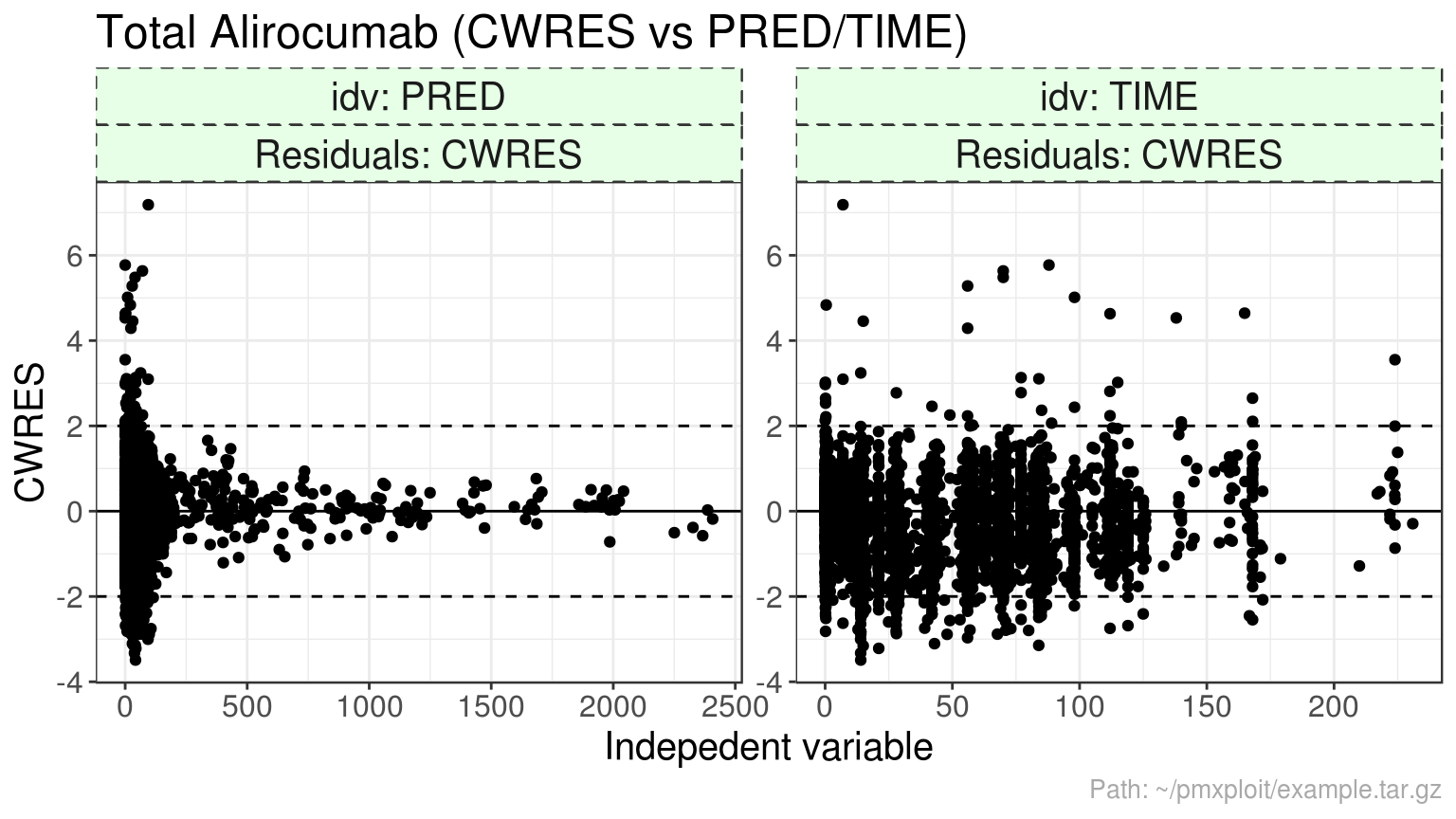
plot_residuals(run, compartment = 2, residuals = c("CWRES"), type = "histogram")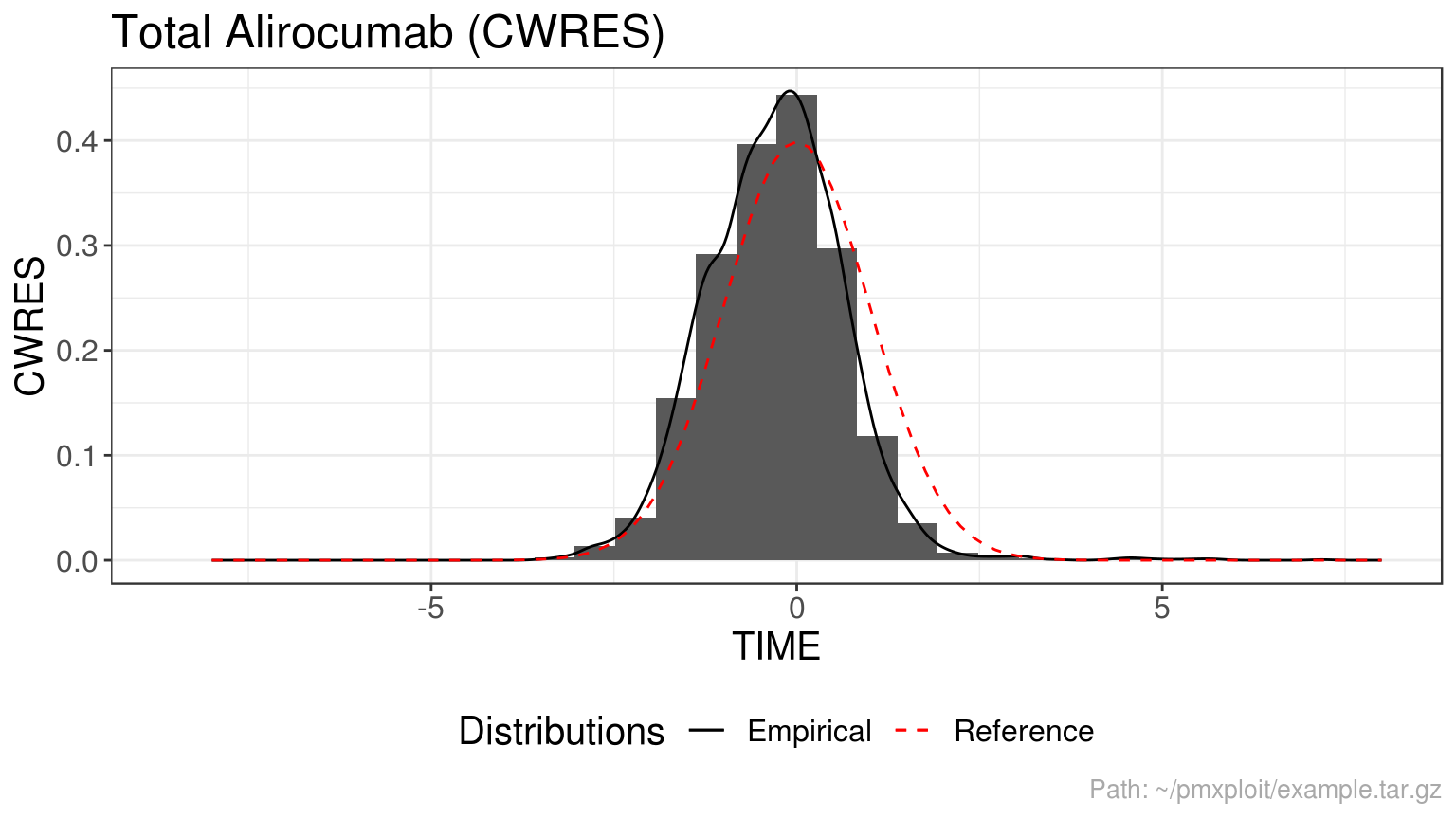
Individual predictions profiles
plot_individual_profiles(run, compartment = 2, ids = 1:4, predictions = c("PRED", "IPRED"), y_scale = "log")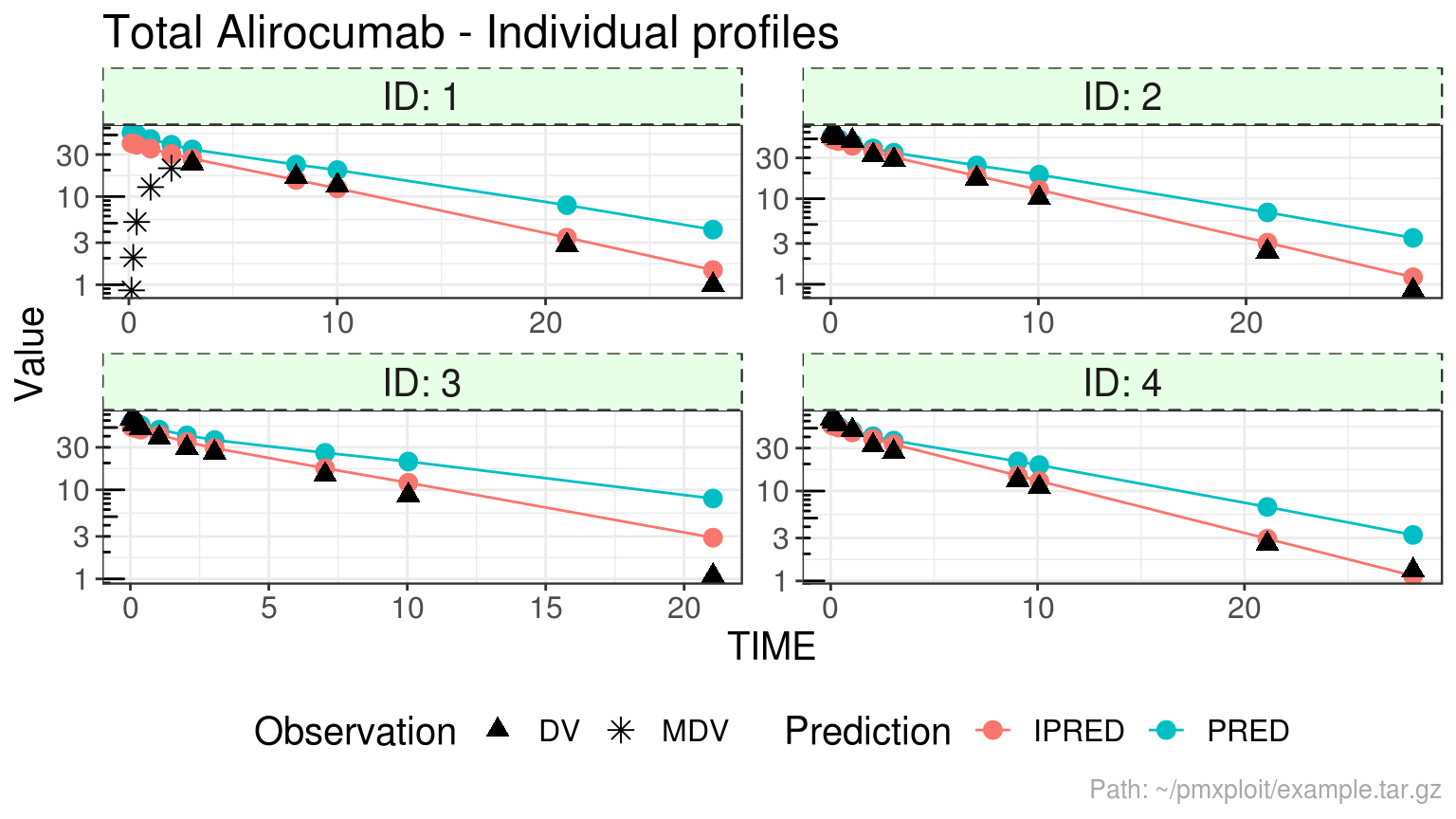
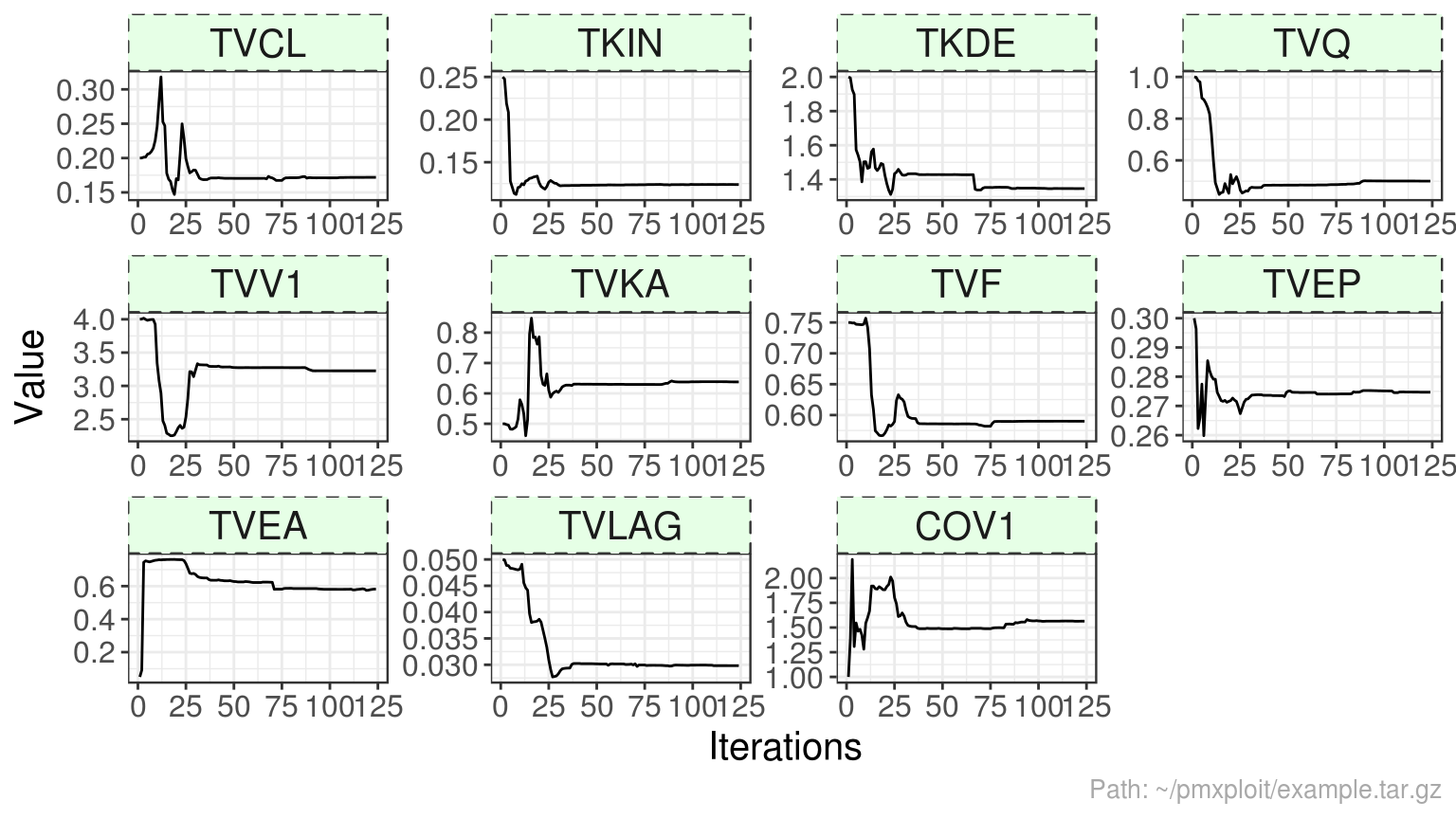
Population parameters
THETA
last_estimation <- last(run$estimations)
last_estimation$thetas %>%
mutate(rse = scales::percent(rse)) %>%
kable()| id | name | estimate | se | rse | ci_low | ci_up |
|---|---|---|---|---|---|---|
| THETA1 | TVCL | 0.1718969 | 0.0152002 | 8.84% | 0.1414965 | 0.2022974 |
| THETA2 | TKON | 559.0000000 | NA | NA% | NA | NA |
| THETA3 | TKIN | 0.1238032 | 0.0022245 | 1.80% | 0.1193541 | 0.1282523 |
| THETA4 | TKDE | 1.3463553 | 0.0375067 | 2.79% | 1.2713418 | 1.4213687 |
| THETA5 | TVQ | 0.5003945 | 0.0285826 | 5.71% | 0.4432292 | 0.5575597 |
| THETA6 | TVV1 | 3.2259046 | 0.2038901 | 6.32% | 2.8181244 | 3.6336848 |
| THETA7 | TVV2 | 2.6120000 | NA | NA% | NA | NA |
| THETA8 | TVKA | 0.6377794 | 0.0467722 | 7.33% | 0.5442350 | 0.7313237 |
| THETA9 | TVF | 0.5900018 | 0.0239372 | 4.06% | 0.5421275 | 0.6378761 |
| THETA10 | TVEP | 0.2746841 | 0.0015050 | 0.55% | 0.2716741 | 0.2776941 |
| THETA11 | TVEA | 0.5817956 | 0.0223812 | 3.85% | 0.5370331 | 0.6265581 |
| THETA12 | TVLAG | 0.0298266 | 0.0010994 | 3.69% | 0.0276277 | 0.0320255 |
| THETA13 | COV1 | 1.5642857 | 0.0850631 | 5.44% | 1.3941594 | 1.7344120 |
OMEGA
# OMEGA matrix
kable(last_estimation$omega_matrix)| ETCL | EKON | EKIN | EKDE | ETQ | ETV1 | ETV2 | ETKA | ETF | |
|---|---|---|---|---|---|---|---|---|---|
| ETCL | 0.279196 | 0 | 0.0000000 | 0.0000000 | 0.0000000 | 0.000000 | 0 | 0.0000000 | 0.0000000 |
| EKON | 0.000000 | 0 | 0.0000000 | 0.0000000 | 0.0000000 | 0.000000 | 0 | 0.0000000 | 0.0000000 |
| EKIN | 0.000000 | 0 | 0.0580908 | 0.0000000 | 0.0000000 | 0.000000 | 0 | 0.0000000 | 0.0000000 |
| EKDE | 0.000000 | 0 | 0.0000000 | 0.1705345 | 0.0000000 | 0.000000 | 0 | 0.0000000 | 0.0000000 |
| ETQ | 0.000000 | 0 | 0.0000000 | 0.0000000 | 0.0691066 | 0.000000 | 0 | 0.0000000 | 0.0000000 |
| ETV1 | 0.000000 | 0 | 0.0000000 | 0.0000000 | 0.0000000 | 0.099037 | 0 | 0.0000000 | 0.0000000 |
| ETV2 | 0.000000 | 0 | 0.0000000 | 0.0000000 | 0.0000000 | 0.000000 | 0 | 0.0000000 | 0.0000000 |
| ETKA | 0.000000 | 0 | 0.0000000 | 0.0000000 | 0.0000000 | 0.000000 | 0 | 0.4644621 | 0.0000000 |
| ETF | 0.000000 | 0 | 0.0000000 | 0.0000000 | 0.0000000 | 0.000000 | 0 | 0.0000000 | 0.2895511 |
# OMEGA matrix as a table, with RSE% and IC 95%
last_estimation$omega %>%
mutate(rse = scales::percent(rse)) %>%
kable()| eta1 | eta2 | estimate | se | rse | ci_low | ci_up | cv |
|---|---|---|---|---|---|---|---|
| ETCL | ETCL | 0.2791960 | 0.0761155 | 27.3% | 0.1300096 | 0.4283823 | 0.5675090 |
| EKIN | EKIN | 0.0580908 | 0.0046239 | 8.0% | 0.0490280 | 0.0671536 | 0.2445633 |
| EKDE | EKDE | 0.1705345 | 0.0164141 | 9.6% | 0.1383628 | 0.2027062 | 0.4312059 |
| ETQ | ETQ | 0.0691066 | 0.0297647 | 43.1% | 0.0107678 | 0.1274454 | 0.2674891 |
| ETV1 | ETV1 | 0.0990370 | 0.0165851 | 16.7% | 0.0665301 | 0.1315438 | 0.3226563 |
| ETKA | ETKA | 0.4644621 | 0.0494723 | 10.7% | 0.3674963 | 0.5614279 | 0.7688681 |
| ETF | ETF | 0.2895511 | 0.0717353 | 24.8% | 0.1489500 | 0.4301522 | 0.5795064 |
Individuals
Parameters
Distributions
summarize_parameters_distributions(run)
#> # A tibble: 11 x 12
#> parameter n n_distinct mean median sd `5.0%` `25.0%`
#> <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 CL 527 493 1.68e-1 1.72e-1 3.05e-2 1.15e-1 1.58e-1
#> 2 KON 527 1 5.59e+2 5.59e+2 0. 5.59e+2 5.59e+2
#> 3 KSS 527 33 5.80e-1 5.80e-1 4.68e-5 5.80e-1 5.80e-1
#> 4 KINT 527 507 1.29e-1 1.25e-1 2.62e-2 9.35e-2 1.13e-1
#> 5 KDEG 527 520 1.38e+0 1.37e+0 4.46e-1 7.78e-1 1.09e+0
#> 6 Q 527 500 5.03e-1 5.00e-1 3.30e-2 4.65e-1 4.92e-1
#> 7 V1 527 523 4.59e+0 4.61e+0 1.30e+0 2.71e+0 3.54e+0
#> 8 V2 527 1 2.61e+0 2.61e+0 0. 2.61e+0 2.61e+0
#> 9 KA 527 496 6.53e-1 6.38e-1 2.52e-1 3.54e-1 5.62e-1
#> 10 ALAG1 527 1 2.98e-2 2.98e-2 0. 2.98e-2 2.98e-2
#> 11 F1 527 494 6.08e-1 5.95e-1 1.20e-1 4.62e-1 5.42e-1
#> # … with 4 more variables: `75.0%` <dbl>, `95.0%` <dbl>, min <dbl>,
#> # max <dbl>
plot_parameters_distributions(run, parameters = "eta")
#> Warning: Computation failed in `stat_bin()`:
#> `binwidth` must be positive
#> Warning: Computation failed in `stat_bin()`:
#> `binwidth` must be positive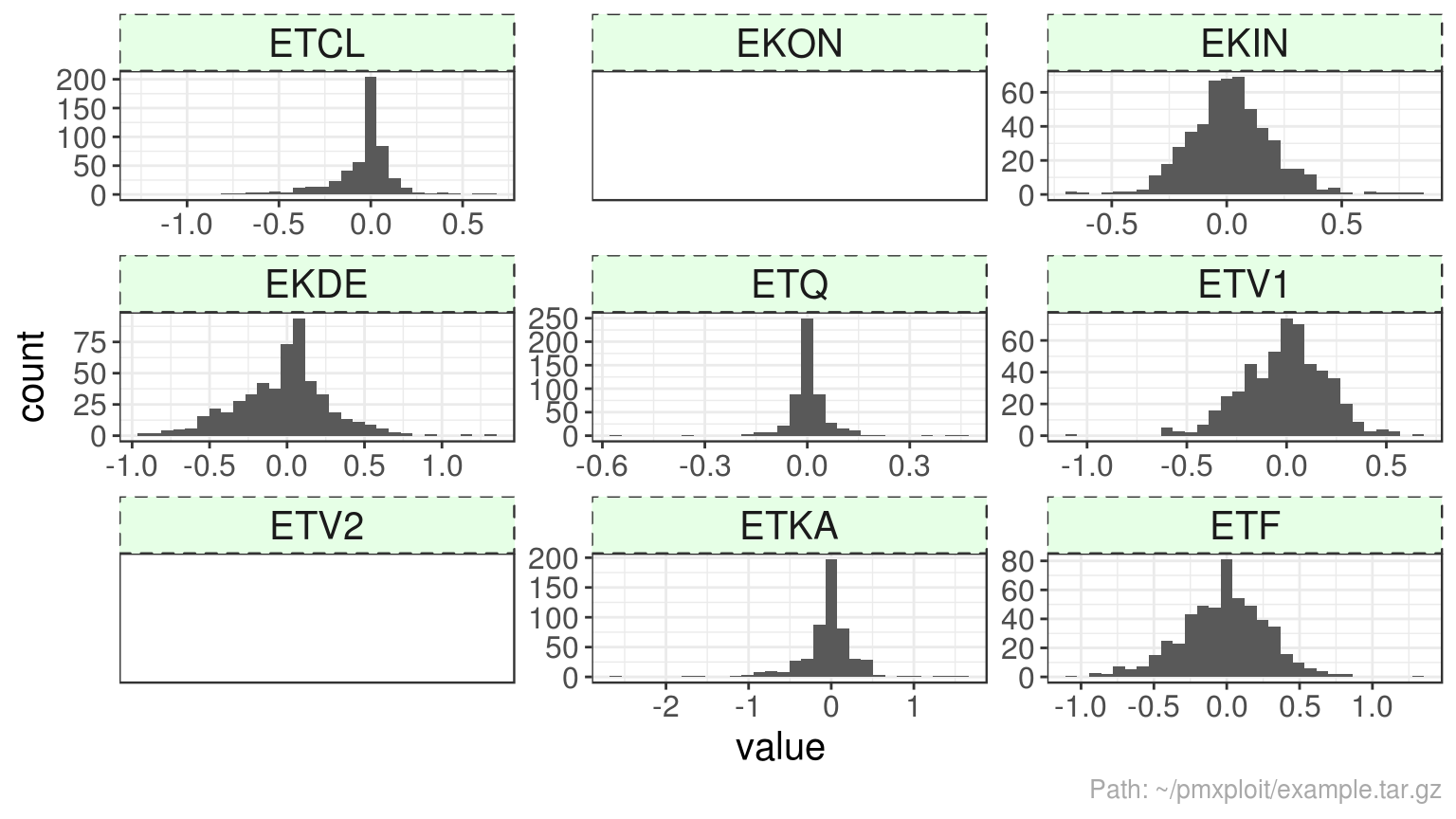
run %>%
group_by(STUD) %>%
plot_parameters_distributions(parameters = c("CL", "V1"), type = "boxplot")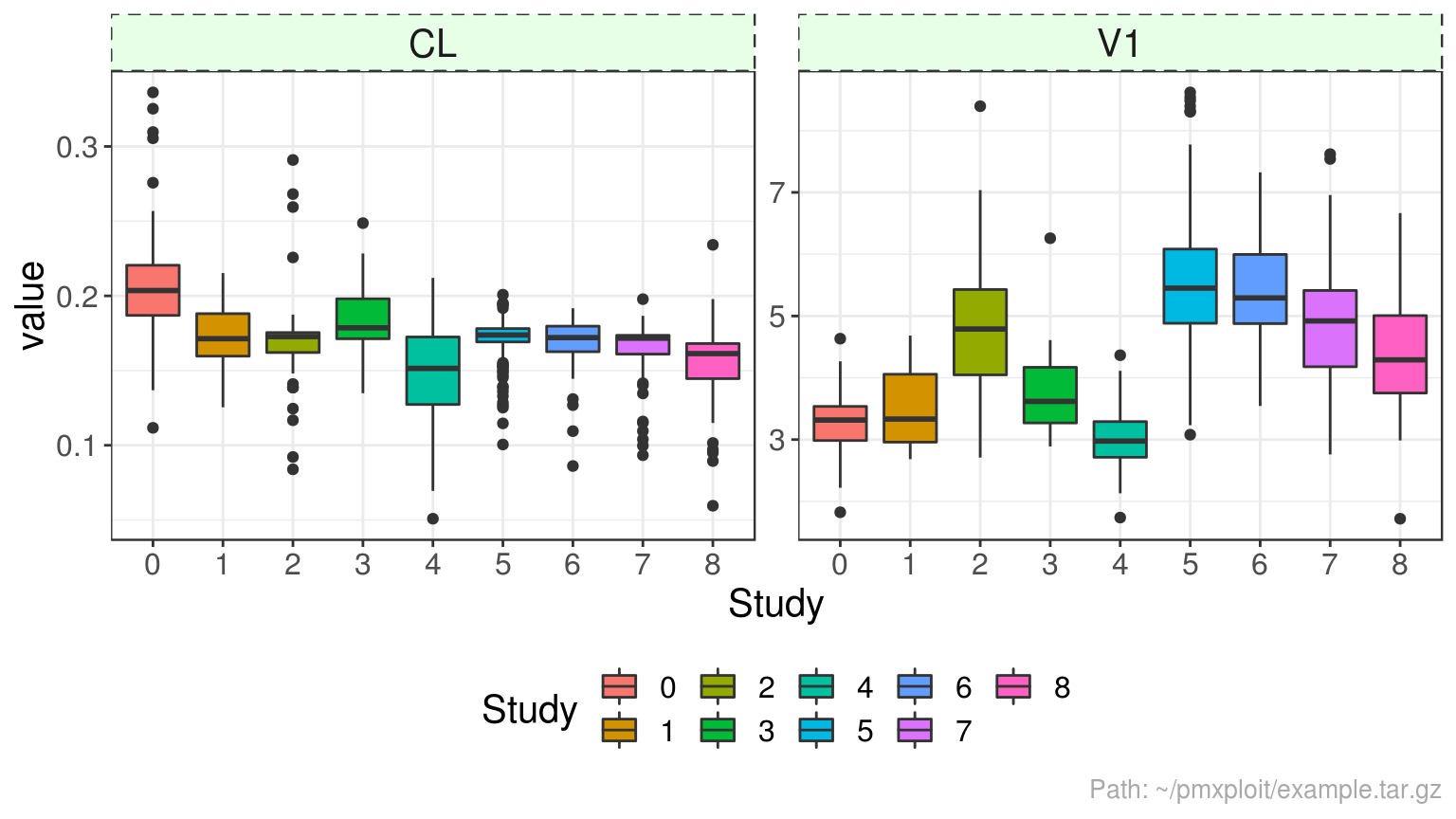
Correlations
summarize_parameters_correlations(run, parameters = "individual")
#> Correlations are not computed for parameters(s) with one unique value: KON, V2, ALAG1
#> V1 F1 Q KSS KINT KA
#> V1 1.00000000 -0.58741001 -0.05479555 0.1106159 0.1098006 0.30323370
#> F1 -0.58741001 1.00000000 0.13141825 0.2147780 0.2165945 -0.06226802
#> Q -0.05479555 0.13141825 1.00000000 0.3817025 0.3792538 0.05201759
#> KSS 0.11061590 0.21477798 0.38170253 1.0000000 0.9981868 0.19737220
#> KINT 0.10980058 0.21659448 0.37925379 0.9981868 1.0000000 0.19826681
#> KA 0.30323370 -0.06226802 0.05201759 0.1973722 0.1982668 1.00000000
#> CL 0.30095621 0.02300692 0.09799981 0.3480292 0.3480398 0.22345707
#> KDEG 0.16623895 -0.04360299 0.09469923 0.2157855 0.2162849 0.17495309
#> CL KDEG
#> V1 0.30095621 0.16623895
#> F1 0.02300692 -0.04360299
#> Q 0.09799981 0.09469923
#> KSS 0.34802922 0.21578546
#> KINT 0.34803981 0.21628493
#> KA 0.22345707 0.17495309
#> CL 1.00000000 0.33390241
#> KDEG 0.33390241 1.00000000
plot_parameters_correlations(run, parameters = "eta", type = "heatmap")
#> Correlations are not computed for parameters(s) with one unique value: EKON, ETV2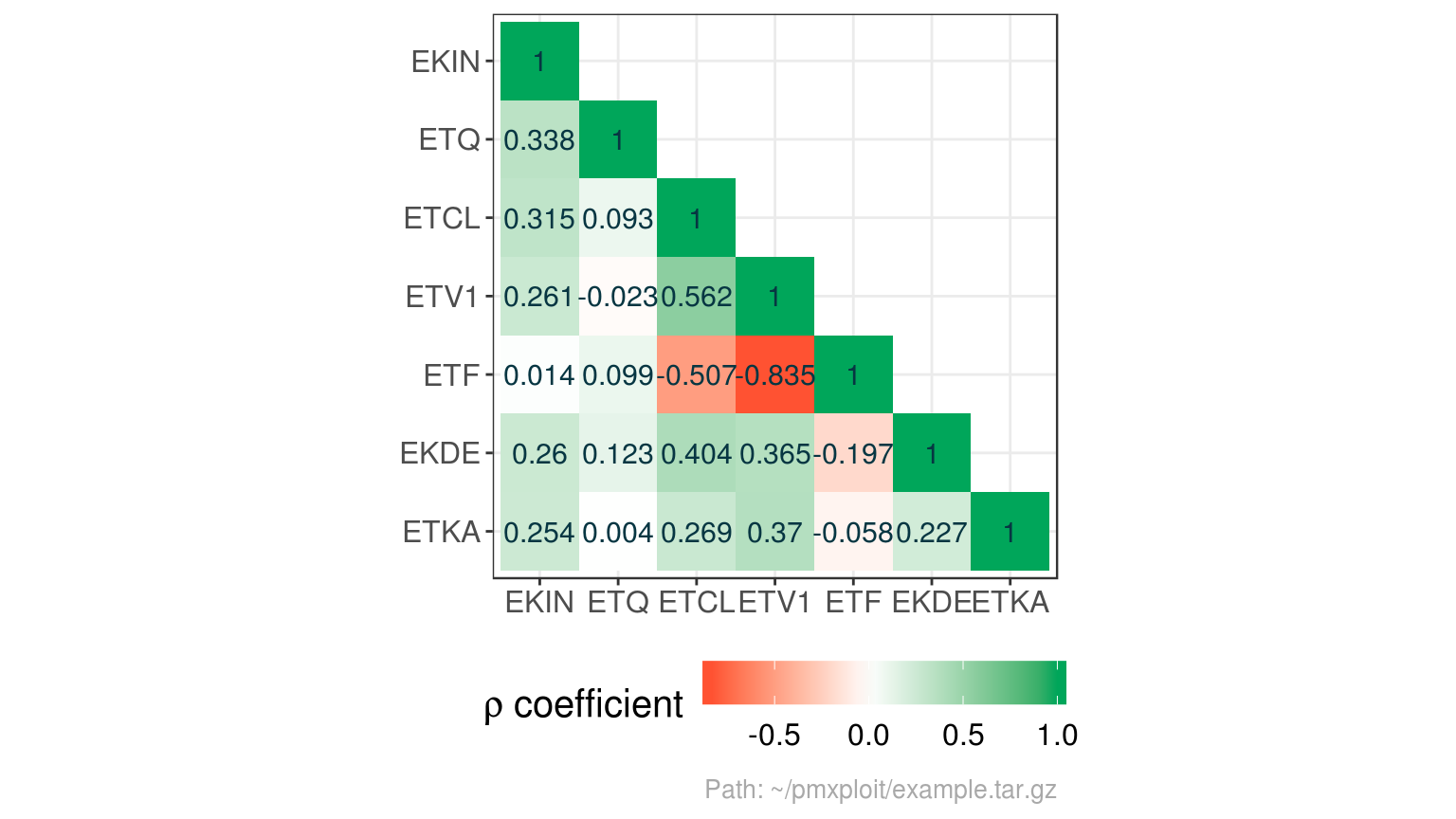
Continuous covariates
Distributions
summarize_continuous_covariates(run)
#> # A tibble: 8 x 12
#> covariate n n_distinct mean median sd `5.0%` `25.0%` `75.0%`
#> <fct> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Age (y) 527 58 52.5 56 13.0 24 46 62
#> 2 BMI (kg/… 527 438 28.0 27.4 4.60 21.5 24.7 30.7
#> 3 Baseline… 527 320 140. 134. 33.0 101 117. 157.
#> 4 Creatini… 527 517 109. 104. 30.4 67.6 87.3 128.
#> 5 Baseline… 527 291 2.55 2.39 1.07 1.22 1.81 3.08
#> 6 Glomerul… 527 480 94.1 90.8 21.1 66.6 79.6 105.
#> 7 Baseline… 527 437 7.66 6.99 3.06 3.73 5.61 9.14
#> 8 Weight (… 527 345 80.6 79.2 16.4 57.3 69.0 89.4
#> # … with 3 more variables: `95.0%` <dbl>, min <dbl>, max <dbl>
plot_continuous_covariates_distributions(run)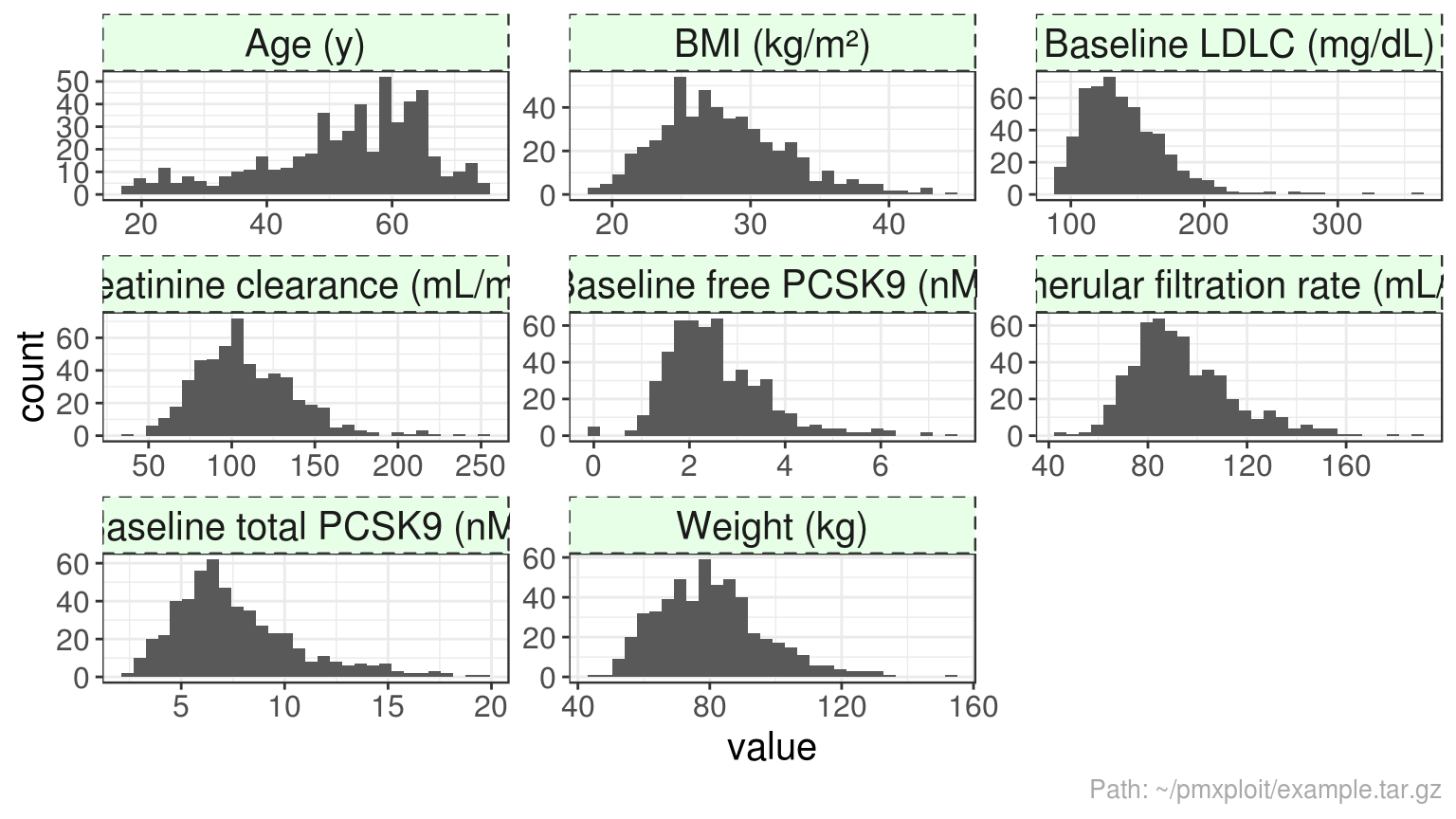
run %>%
group_by(STUD) %>%
plot_continuous_covariates_distributions(type = "boxplot")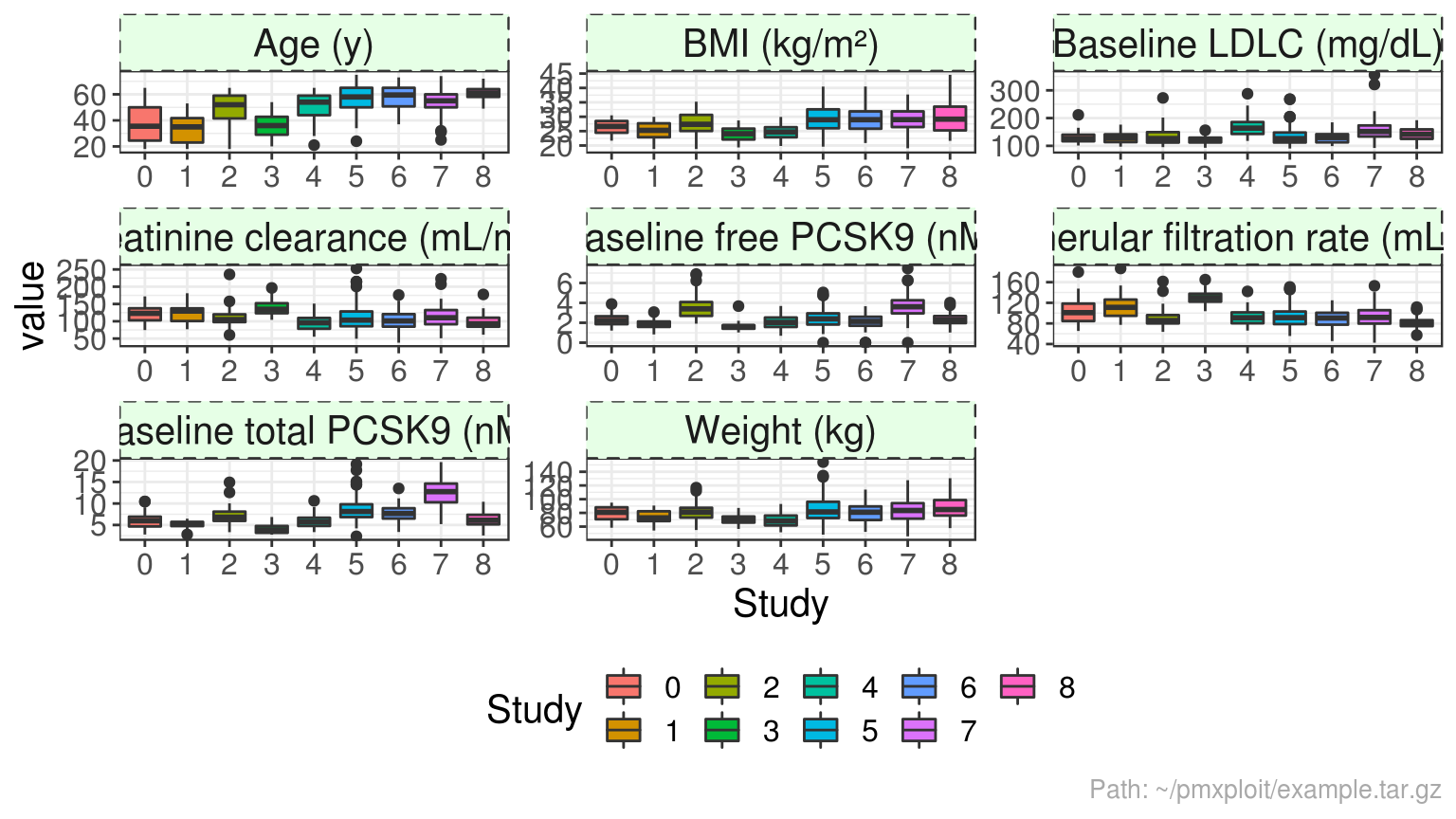
Correlations
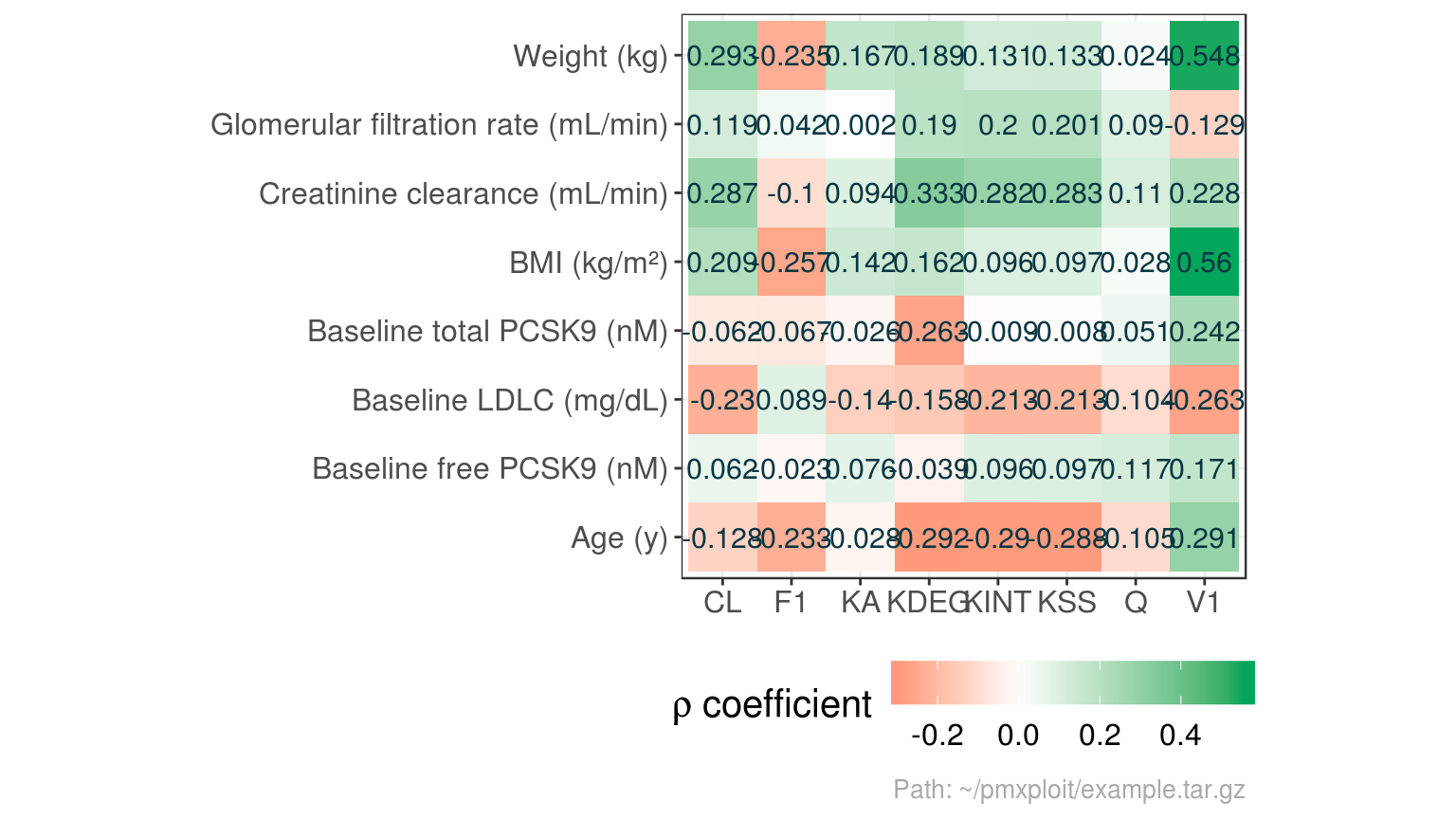
summarize_covariates_correlations(run)
#> Baseline free PCSK9 (nM)
#> Baseline free PCSK9 (nM) 1.00000000
#> Baseline total PCSK9 (nM) 0.63474111
#> Creatinine clearance (mL/min) -0.03390245
#> Age (y) 0.11602376
#> Glomerular filtration rate (mL/min) -0.13572416
#> Baseline LDLC (mg/dL) -0.05890704
#> BMI (kg/m²) 0.14352343
#> Weight (kg) 0.10401265
#> Baseline total PCSK9 (nM)
#> Baseline free PCSK9 (nM) 0.63474111
#> Baseline total PCSK9 (nM) 1.00000000
#> Creatinine clearance (mL/min) -0.06800086
#> Age (y) 0.26465204
#> Glomerular filtration rate (mL/min) -0.14715975
#> Baseline LDLC (mg/dL) 0.12308252
#> BMI (kg/m²) 0.17724420
#> Weight (kg) 0.08670552
#> Creatinine clearance (mL/min)
#> Baseline free PCSK9 (nM) -0.03390245
#> Baseline total PCSK9 (nM) -0.06800086
#> Creatinine clearance (mL/min) 1.00000000
#> Age (y) -0.52845720
#> Glomerular filtration rate (mL/min) 0.67538184
#> Baseline LDLC (mg/dL) -0.12739227
#> BMI (kg/m²) 0.40805133
#> Weight (kg) 0.56597426
#> Age (y)
#> Baseline free PCSK9 (nM) 0.11602376
#> Baseline total PCSK9 (nM) 0.26465204
#> Creatinine clearance (mL/min) -0.52845720
#> Age (y) 1.00000000
#> Glomerular filtration rate (mL/min) -0.51708487
#> Baseline LDLC (mg/dL) 0.01378944
#> BMI (kg/m²) 0.20298742
#> Weight (kg) 0.06328087
#> Glomerular filtration rate (mL/min)
#> Baseline free PCSK9 (nM) -0.13572416
#> Baseline total PCSK9 (nM) -0.14715975
#> Creatinine clearance (mL/min) 0.67538184
#> Age (y) -0.51708487
#> Glomerular filtration rate (mL/min) 1.00000000
#> Baseline LDLC (mg/dL) -0.03312037
#> BMI (kg/m²) -0.15911903
#> Weight (kg) -0.10956357
#> Baseline LDLC (mg/dL) BMI (kg/m²)
#> Baseline free PCSK9 (nM) -0.05890704 0.1435234
#> Baseline total PCSK9 (nM) 0.12308252 0.1772442
#> Creatinine clearance (mL/min) -0.12739227 0.4080513
#> Age (y) 0.01378944 0.2029874
#> Glomerular filtration rate (mL/min) -0.03312037 -0.1591190
#> Baseline LDLC (mg/dL) 1.00000000 -0.1510291
#> BMI (kg/m²) -0.15102914 1.0000000
#> Weight (kg) -0.16168067 0.7920918
#> Weight (kg)
#> Baseline free PCSK9 (nM) 0.10401265
#> Baseline total PCSK9 (nM) 0.08670552
#> Creatinine clearance (mL/min) 0.56597426
#> Age (y) 0.06328087
#> Glomerular filtration rate (mL/min) -0.10956357
#> Baseline LDLC (mg/dL) -0.16168067
#> BMI (kg/m²) 0.79209179
#> Weight (kg) 1.00000000
plot_covariates_correlations(run, type = "heatmap")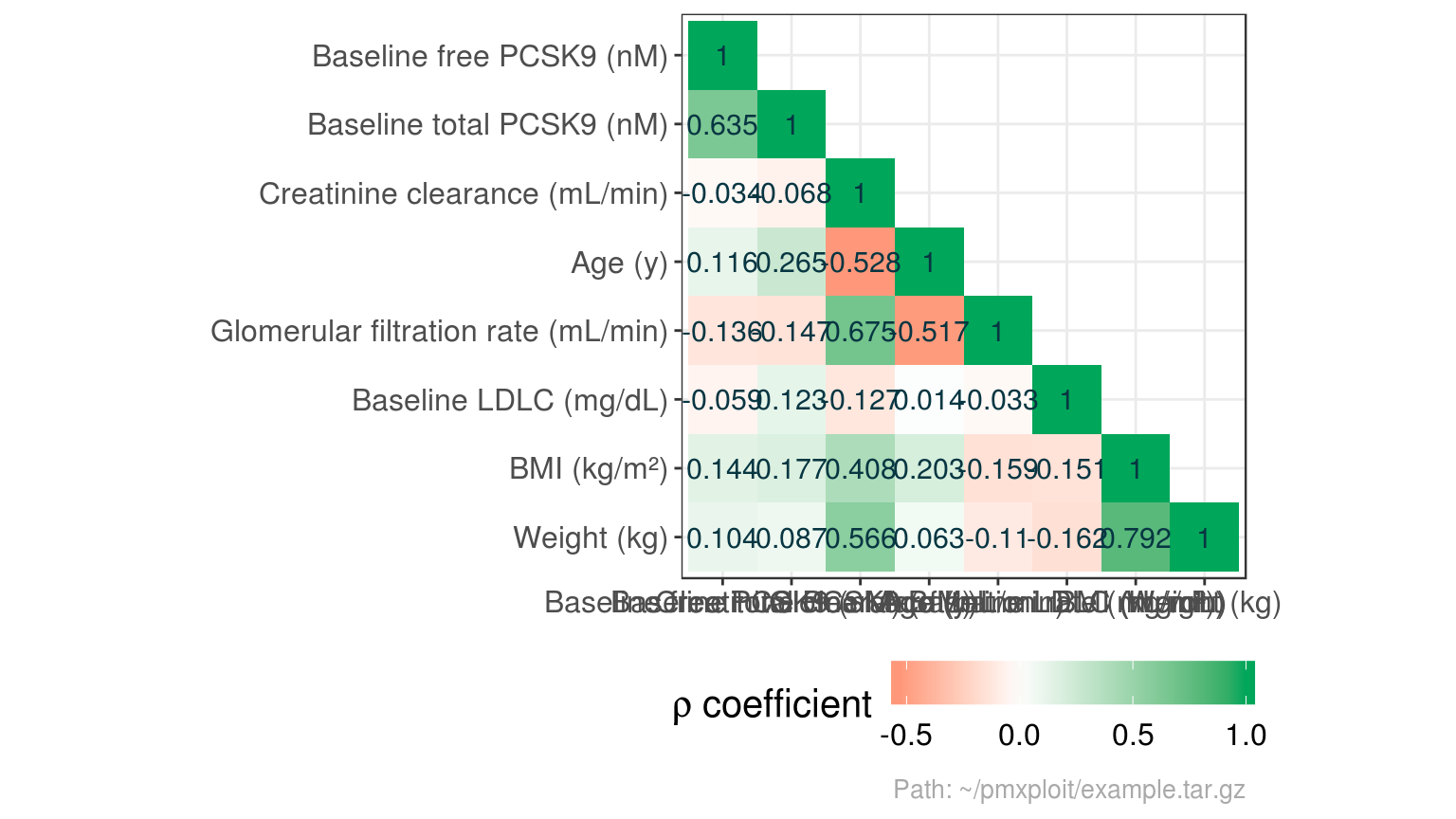
run %>%
group_by(STUD) %>%
plot_covariates_correlations(type = "scatterplot")
#> Warning in GGally::ggscatmat(as.data.frame(select(df, -ID))): Factor
#> variables are omitted in plot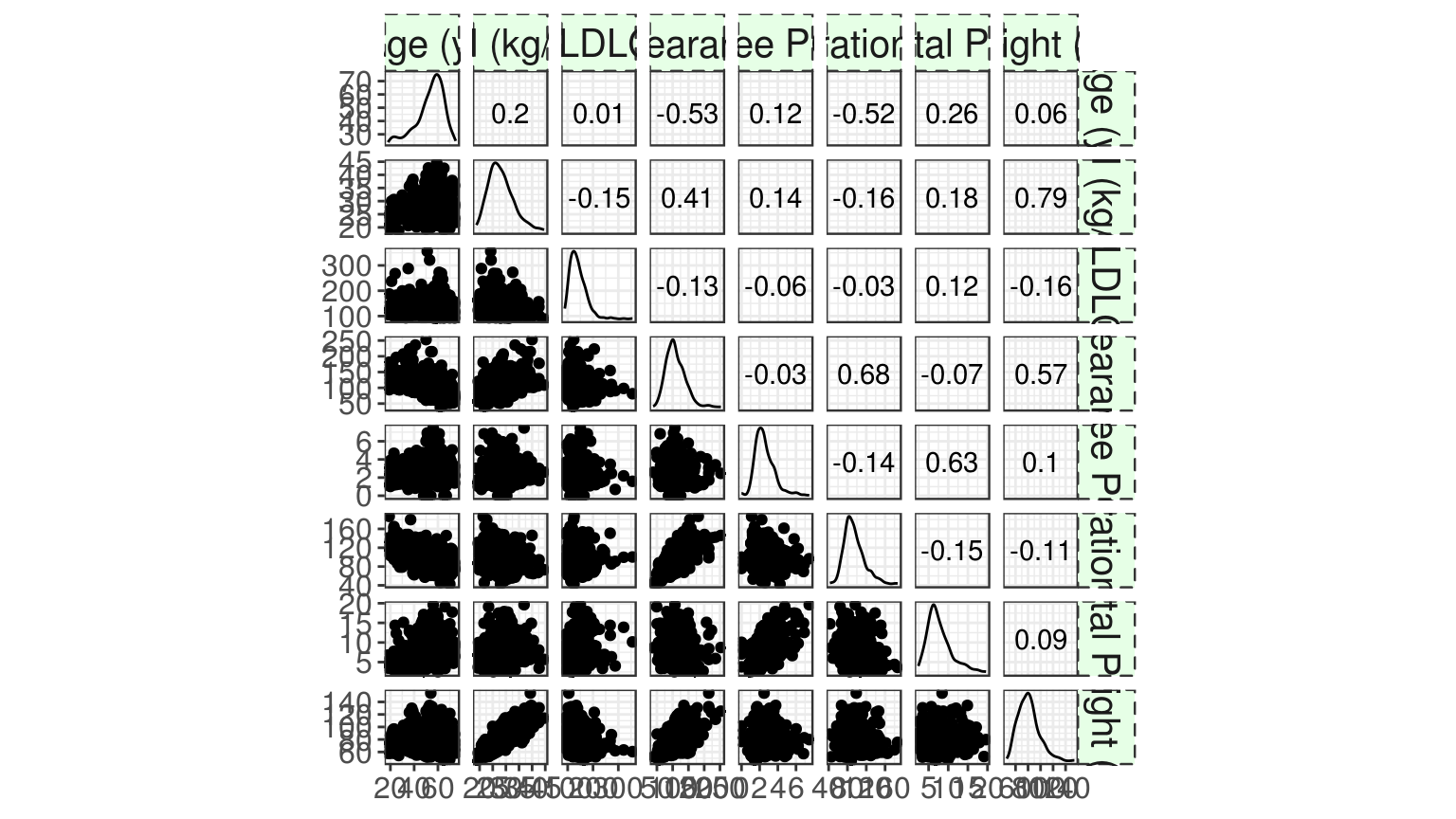
Quality criteria
# Overall QC
qc <- quality_criteria(run, predictions = "PRED")
# QC by study
qc_stud <- run %>%
group_by(STUD) %>%
quality_criteria(predictions = "PRED")
kable(qc$standard)
|
qc_stud %>%
select(Study, standard) %>%
unnest() %>%
kable()| Study | max_err | aafe |
|---|---|---|
| 0 | 786.5671 | 1.402490 |
| 1 | 120.9971 | 1.487727 |
| 2 | 171.3205 | 1.516194 |
| 3 | 113.7925 | 1.569381 |
| 4 | 460.8415 | 1.550188 |
| 5 | 190.3589 | 1.605442 |
| 6 | 117.1923 | 1.637036 |
| 7 | 228.3282 | 1.624440 |
| 8 | 146.0201 | 1.847918 |
# Bias
qc_stud %>%
select(Study, bias) %>%
unnest() %>%
mutate(relative_value = scales::percent(relative_value)) %>%
rename(`Mean Prediction Error (%)` = relative_value) %>%
kable()| Study | value | ci_low | ci_up | Mean Prediction Error (%) |
|---|---|---|---|---|
| 0 | -14.2811313 | -22.301736 | -6.260527 | -5.8% |
| 1 | 0.4946943 | -1.185524 | 2.174913 | 1.8% |
| 2 | -4.1641145 | -4.904954 | -3.423276 | -18.4% |
| 3 | 8.4579500 | 6.735203 | 10.180697 | 25.3% |
| 4 | -10.8373662 | -12.183189 | -9.491543 | -22.2% |
| 5 | 1.9028940 | 1.002670 | 2.803118 | 5.9% |
| 6 | -0.3312528 | -2.009636 | 1.347131 | -0.9% |
| 7 | -9.5851697 | -11.691082 | -7.479257 | -20.6% |
| 8 | -13.3829373 | -15.581440 | -11.184434 | -31.6% |
# Precision
qc_stud %>%
select(Study, precision) %>%
unnest() %>%
mutate(relative_value = scales::percent(relative_value)) %>%
rename(`Root Mean Square Error (%)` = relative_value) %>%
kable()| Study | value | ci_low | ci_up | Root Mean Square Error (%) |
|---|---|---|---|---|
| 0 | 119.60062 | 101.32976 | 135.42856 | 48.9% |
| 1 | 19.36620 | 16.31571 | 21.99766 | 71.3% |
| 2 | 16.07381 | 14.11605 | 17.81773 | 71.2% |
| 3 | 22.88012 | 20.21317 | 25.26712 | 68.4% |
| 4 | 30.11989 | 24.73595 | 34.67777 | 61.7% |
| 5 | 20.95346 | 19.33192 | 22.45822 | 65.5% |
| 6 | 21.78440 | 19.38433 | 23.94509 | 56.7% |
| 7 | 33.23176 | 28.39221 | 37.45106 | 71.5% |
| 8 | 28.31826 | 24.68632 | 31.53464 | 66.8% |